Content from Writing Reproducible Python
Last updated on 2026-02-10 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What is the difference between the standard library and third-party packages?
- How do I share a script so that it runs on someone else’s computer?
Objectives
- Import and use the
pathlibstandard library. - Identify when a script requires external dependencies (like
numpy). - Write a self-contained script that declares its own dependencies using inline metadata.
- Share a script which reproducibly handles
condadependencies alongside Python.
The Humble Script
Most research software starts as a single file. You have some data, you need to analyze it, and you write a sequence of commands to get the job done.
Let’s start by creating a script that generates some data and saves
it. We will use the standard library module pathlib to handle file paths safely across
operating systems (Windows/macOS/Linux).
PYTHON
import random
from pathlib import Path
# Define output directory
DATA_DIR = Path("data")
DATA_DIR.mkdir(exist_ok=True)
def generate_trajectory(n_steps=100):
print(f"Generating trajectory with {n_steps} steps...")
path = [0.0]
for _ in range(n_steps):
# Random walk step
step = random.uniform(-0.5, 0.5)
path.append(path[-1] + step)
return path
if __name__ == "__main__":
traj = generate_trajectory()
output_file = DATA_DIR / "trajectory.txt"
with open(output_file, "w") as f:
for point in traj:
f.write(f"{point}\n")
print(f"Saved to {output_file}")This script uses only Built-in modules (random, pathlib).
You can send this file to anyone with Python installed, and it will
run.
The Need for External Libraries
Standard Python is powerful, but for scientific work, we almost
always need the “Scientific Stack”: numpy,
pandas/polars, or matplotlib.
Let’s modify our script to calculate statistics using numpy.
PYTHON
import random
from pathlib import Path
import numpy as np # new dependency!!
DATA_DIR = Path("data")
DATA_DIR.mkdir(exist_ok=True)
def generate_trajectory(n_steps=100):
# Use numpy for efficient array generation
steps = np.random.uniform(-0.5, 0.5, n_steps)
trajectory = np.cumsum(steps)
return trajectory
if __name__ == "__main__":
traj = generate_trajectory()
print(f"Mean position: {np.mean(traj):.4f}")
print(f"Std Dev: {np.std(traj):.4f}")The Dependency Problem
If you send this updated file to a colleague who just installed Python, what happens when they run it?
The Modern Solution: PEP 723 Metadata
Traditionally, you would send a requirements.txt file alongside your script, or
leave comments in the script, or try to add documentation in an
email.
But files get separated, and versions get desynchronized.
PEP
723 is a Python standard that allows you to embed
dependency information directly into the script file. Tools like uv (a fast Python package manager) can read this
header and automatically set up the environment for you.
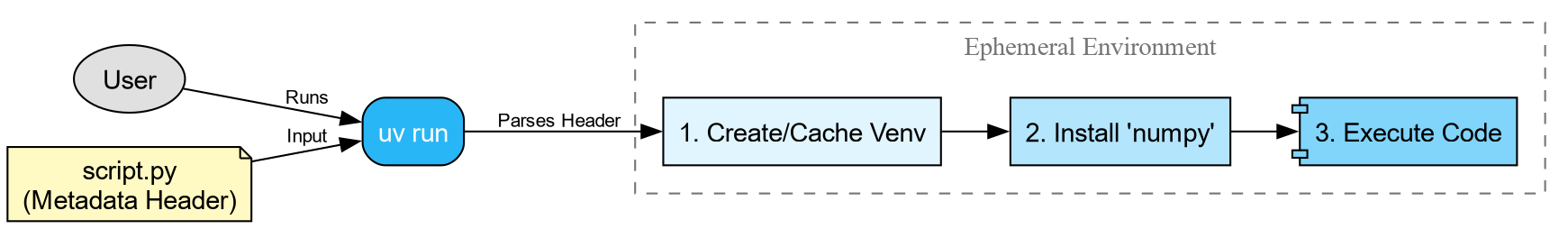
We can add a special comment block at the top of our script:
PYTHON
# /// script
# requires-python = ">=3.11"
# dependencies = [
# "numpy",
# ]
# ///
import numpy as np
print("Hello I don't crash anymore..")
# ... rest of script ...Now, instead of manually installing numpy, you run the script using uv:
When you run this command:
-
uvreads the metadata block. - It creates a temporary, isolated virtual environment.
- It installs the specified version of
numpy. - It executes the script.
This guarantees that anyone with uv
installed can run your script immediately, without messing up their own
python environments.
Beyond Python: The pixibang
PEP 723 is fantastic for installable Python packages [fn:: most often this means things you can find on PyPI).
However, for scientific software, we often rely on compiled binaries
and libraries that are not Python packages—things like LAMMPS, GROMACS,
or eOn a server-client
tool for exploring the potential energy surfaces of atomistic
systems.
If your script needs to run a C++ binary, pip and uv cannot
help you easily. This is where pixi comes in.
pixi is a package manager
built on the conda ecosystem. It can
install Python packages and compiled binaries. We can
use a “pixibang”
script to effectively replicate the PEP 723 experience, but for the
entire system stack.
Example: Running minimizations with eOn and PET-MAD
Let’s write a script that drives a geometry minimization 1. This requires:
- Metatrain/Torch
- For the machine learning potential.
- rgpycrumbs
- For helper utilities.
- eOn Client
- The compiled C++ binary that actually performs the minimization.
First, we need to create the input geometry file pos.con in our directory:
BASH
cat << 'EOF' > pos.con
Generated by ASE
preBox_header_2
25.00 25.00 25.00
90.00 90.00 90.00
postBox_header_1
postBox_header_2
4
2 1 2 4
12.01 16.00 14.01 1.01
C
Coordinates of Component 1
11.04 11.77 12.50 0 0
12.03 10.88 12.50 0 1
O
Coordinates of Component 2
14.41 13.15 12.44 0 2
N
Coordinates of Component 3
13.44 13.86 12.46 0 3
12.50 14.51 12.49 0 4
H
Coordinates of Component 4
10.64 12.19 13.43 0 5
10.59 12.14 11.58 0 6
12.49 10.52 13.42 0 7
12.45 10.49 11.57 0 8
EOFNow, create the script eon_min.py. Note
the shebang line!
PYTHON
#!/usr/bin/env -S pixi exec --spec eon --spec uv -- uv run
# /// script
# requires-python = ">=3.11"
# dependencies = [
# "ase",
# "metatrain",
# "rgpycrumbs",
# ]
# ///
from pathlib import Path
import subprocess
from rgpycrumbs.eon.helpers import write_eon_config
from rgpycrumbs.run.jupyter import run_command_or_exit
repo_id = "lab-cosmo/upet"
tag = "v1.1.0"
url_path = f"models/pet-mad-s-{tag}.ckpt"
fname = Path(url_path.replace(".ckpt", ".pt"))
url = f"https://huggingface.co/{repo_id}/resolve/main/{url_path}"
fname.parent.mkdir(parents=True, exist_ok=True)
subprocess.run(
[
"mtt",
"export",
url,
"-o",
fname,
],
check=True,
)
print(f"Successfully exported {fname}.")
min_settings = {
"Main": {"job": "minimization", "random_seed": 706253457},
"Potential": {"potential": "Metatomic"},
"Metatomic": {"model_path": fname.absolute()},
"Optimizer": {
"max_iterations": 2000,
"opt_method": "lbfgs",
"max_move": 0.5,
"converged_force": 0.01,
},
}
write_eon_config(".", min_settings)
run_command_or_exit(["eonclient"], capture=True, timeout=300)Make it executable and run it:
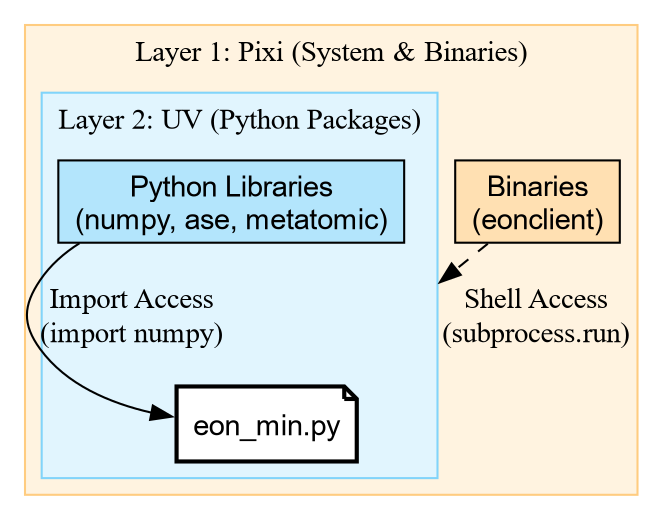
Unpacking the Shebang
The magic happens in this line: #!/usr/bin/env -S pixi exec --spec eon --spec uv -- uv run
This is a chain of tools:
-
pixi exec: Create an environment with
pixi. -
–spec eon: Explicitly request the
eonpackage (which contains the binaryeonclient). -
–spec uv: Explicitly request
uv. -
– uv run: Once the outer environment exists with
eOnanduv, it hands control over touv run. -
PEP 723:
uv runreads the script comments and installs the Python libraries (ase,rgpycrumbs).
This gives us the best of both worlds: pixi provides the
compiled binaries, and uv handles the fast Python
resolution.
The Result
When executed, the script downloads the model, exports it using metatrain, configures eOn, and runs the
binary.
[INFO] - Using best model from epoch None
[INFO] - Model exported to '.../models/pet-mad-s-v1.1.0.pt'
Successfully exported models/pet-mad-s-v1.1.0.pt.
Wrote eOn config to 'config.ini'
EON Client
VERSION: 01e09a5
...
[Matter] 0 0.00000e+00 1.30863e+00 -53.90300
[Matter] 1 1.46767e-02 6.40732e-01 -53.91548
...
[Matter] 51 1.56025e-03 9.85039e-03 -54.04262
Minimization converged within tolerence
Saving result to min.con
Final Energy: -54.04261779785156Challenge: The Pure Python Minimization
Create a script named ase_min.py that
performs the exact same minimization on pos.con, but uses the atomic simulation environment (ASE)
built-in LBFGS optimizer instead of
eOn.
Requirements:
- Do we need
pixi? Try using theuvshebang only (nopixi). - Reuse the model file we exported earlier (
models/pet-mad-s-v1.1.0.pt). - Compare the “User Time” of this script vs the EON script.
Hint: You will need the metatomic package to load the potential in
ASE.
PYTHON
# /// script
# requires-python = ">=3.11"
# dependencies = [
# "ase",
# "metatomic",
# "numpy",
# ]
# ///
from ase.io import read
from ase.optimize import LBFGS
from metatomic.torch.ase_calculator import MetatomicCalculator
def run_ase_min():
atoms = read("pos.con")
# Reuse the .pt file exported by the previous script
atoms.calc = MetatomicCalculator(
"models/pet-mad-s-v1.1.0.pt",
device="cpu"
)
# Setup Optimizer
print(f"Initial Energy: {atoms.get_potential_energy():.5f} eV")
opt = LBFGS(atoms, logfile="-") # Log to stdout
opt.run(fmax=0.01)
print(f"Final Energy: {atoms.get_potential_energy():.5f} eV")
if __name__ == "__main__":
run_ase_min()Initial Energy: -53.90300 eV
Step Time Energy fmax
....
LBFGS: 64 20:42:09 -54.042595 0.017080
LBFGS: 65 20:42:09 -54.042610 0.009133
Final Energy: -54.04261 eVSo we get the same result, but with more steps…
| Feature | EON Script (Pixi) | ASE Script (UV) |
| Shebang | pixi exec ... -- uv run |
uv run |
| Engine | C++ Binary (eonclient) |
Python Loop (LBFGS) |
| Dependencies | System + Python | Pure Python |
| Use Case | HPC / Heavy Simulations | Analysis / Prototyping |
While the Python version seems easier to setup, the eOn C++ client is often more performant, and equally trivial with the c.
- PEP 723 allows inline metadata for Python dependencies.
- Use uv to run single-file scripts with pure Python
requirements (
numpy,pandas). - Use Pixi when your script depends on system
libraries or compiled binaries (
eonclient,ffmpeg). - Combine them with a Pixibang (
pixi exec ... -- uv run) for fully reproducible, complex scientific workflows.
A subset of the Cookbook recipe for saddle point optimization↩︎
Content from Modules, Packages, and The Search Path
Last updated on 2026-02-10 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- How does Python know where to find the libraries you import?
- What distinguishes a python “script” from a python “package”?
- What is an
__init__.pyfile?
Objectives
- Inspect the
sys.pathvariable to understand import resolution. - Differentiate between built-in modules, installed packages, and local code.
- Create a minimal local package structure.
From Scripts to Reusable Code
You have likely written Python scripts before: single files ending in
.py that perform a specific analysis or
task. While scripts are excellent for execution
(running a calculation once), they are often poor at facilitating
reuse.
Imagine you wrote a useful function to calculate the center of mass
of a molecule in analysis.py. A month
later, you start a new project and need that same function. You have two
options:
-
Copy and Paste: You copy the function into your new
script.
- Problem: If you find a bug in the original function, you have to remember to fix it in every copy you made.
- Importing: You tell Python to load the code from the original file.
Option 2 is the foundation of Python packaging. To do this effectively, we must first understand how Python finds the code you ask for.
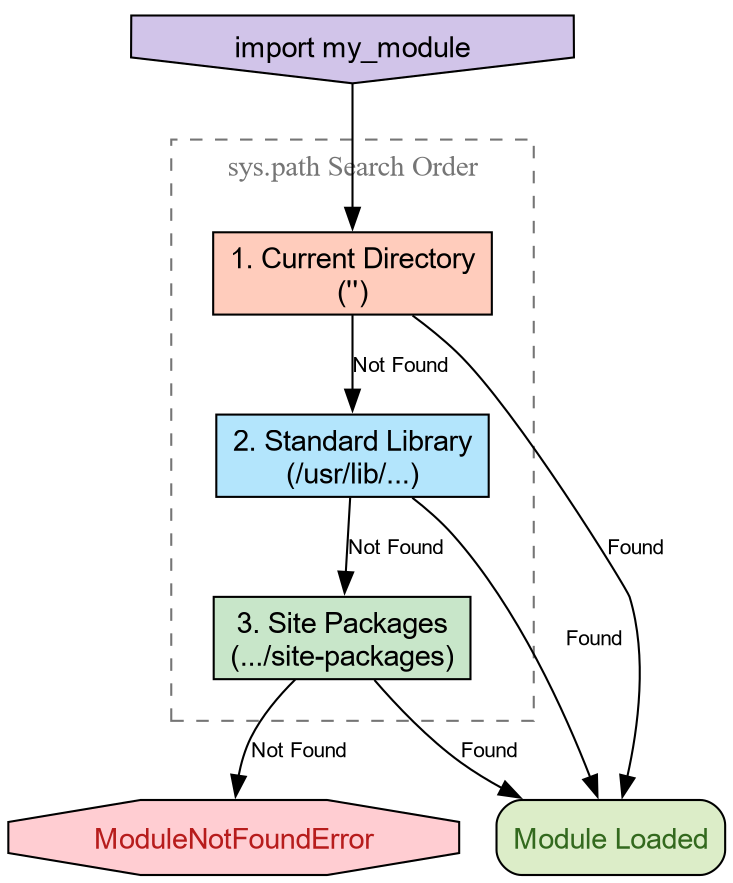
How Python Finds Code
When you type import numpy, Python does
not magically know where that code lives. It follows a deterministic
search procedure. We can see this procedure in action using the built-in
sys module.
PYTHON
['',
'/usr/lib/python314.zip',
'/usr/lib/python3.14',
'/usr/lib/python3.14/lib-dynload',
'/usr/lib/python3.14/site-packages']The variable sys.path is a list of
directory strings. When you import a module, Python scans these
directories in order. The first match wins.
-
The Empty String (’’): This represents the
current working directory. This is why you can always
import a
helper.pyfile if it is sitting right next to your script. -
Standard Library: Locations like
/usr/lib/python3.*contain built-ins likeos,math, andpathlib. -
Site Packages: Directories like
site-packagesordist-packagesare where tools likepip,conda, orpixiplace third-party libraries.
Python will import your local file instead of the
standard library math module.
Why? Because the current working directory
(represented by '' in sys.path) is usually at the top of the list. It
finds your math.py before scanning the
standard library paths. This is called “Shadowing” and is a common
source of bugs!
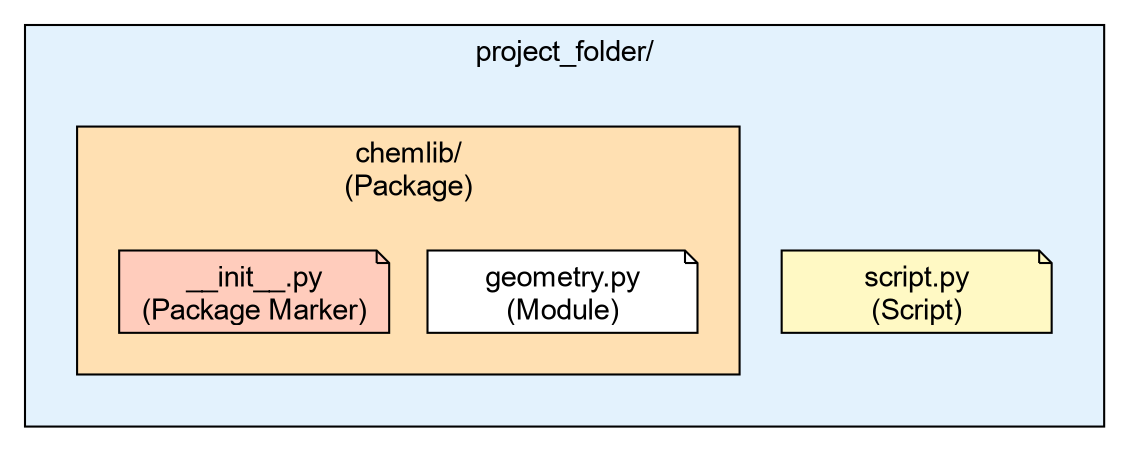
The Anatomy of a Package
- A Module
-
Is simply a single file ending in
.py. - A Package
-
Is a directory containing modules and a special file:
__init__.py.
Let’s create a very simple local package to handle some basic
chemistry geometry. We will call it chemlib.
Now, create a module inside this directory called geometry.py:
Your directory structure should look like this:
project_folder/
├── script.py
└── chemlib/
├── __init__.py
└── geometry.pyThe Role of __init__.py
The __init__.py file tells Python:
“Treat this directory as a package.” It is the first file executed when
you import the package. It can be empty, but it is often used to expose
functions to the top level.
Open `chemlib/_init__.py` and add:
Now, from the project_folder (the
parent directory), launch Python:
Loading chemlib package...
Calculating Center of Mass...
[0.0, 0.0, 0.0]The “It Works on My Machine” Problem
We have created a package, but it is fragile. It relies entirely on
the Current Working Directory being in sys.path.
Challenge: Moving Directories
- Exit your python session.
- Change your directory to go one level up (outside your project
folder):
cd .. - Start Python and try to run
import chemlib.
What happens and why?
Output:
ModuleNotFoundError: No module named 'chemlib'Reason: You moved out of the folder containing chemlib. Since the package is not installed in
the global site-packages, and the current
directory no longer contains it, Python’s search through sys.path fails to find it.
To solve this, we need a standard way to tell Python “This package exists, please add it to your search path permanently.” This is the job of Packaging and Installation.
-
sys.pathis the list of directories Python searches for imports. - The order of search is: Current Directory -> Standard Library -> Installed Packages.
- A Package is a directory containing an
__init__.pyfile. - Code that works locally because of the current directory will fail when shared unless properly installed.
